using peptide surfaces to study the reprogramming of malignant melanoma cells towards identification of a cancer stem cell state
w. jonas reger, kristopher a. kilian
university of illinois urbana-champaign | american cancer society
abstract
the microenvironment that surrounds cells contains many different components of molecules such as proteins and growth factors that influence cellular behavior. by creating microenvironments to influence cells, it will allow us to use materials to control cellular behaviors for biomedical applications in cancer diagnosis and treatment. in this project, we used solid phase peptide synthesis to create short peptides derived from adhesion and signaling proteins. we employed a method using copper-catalyzed azide-alkyne cycloaddition on surfaces to arrange the peptides in self-assembled monolayers (sam) to see how mouse melanoma cells would react to different combinations of peptides and growth factors in terms of marker expressions. adhesion peptides and combinations of cancer implicated growth factors (atwlppr, htsdqtn, cpqprpl, and svvyglr) were immobilized onto gold-coated coverslips. b16f10 melanoma cells were later seeded and stained for markers of the cellular epigenetic state and pluripotency, acetylated lysine and nanog respectively. peptides involved in epidermal growth factor signaling increased lysine acetylation in adherent b16f10s compared to the adhesion peptide alone. this platform will prove useful for the discovery of synthetic surfaces that control melanoma epigenetics and regulate gene expression for studying oncogenesis and reprogramming melanoma to a cancer stem cell state.
1 introduction
melanoma stem cells are a putative cancer stem cell that has proven difficult to identify. one difficulty with their identification as stem cells is that they are difficult to isolate. a potential promising strategy for the in vitro derivation of melanoma stem cells is reprogramming by forced expression of transcription factors (yamanaka, cell 2006). in order to reprogram melanoma cells, they need to de-differentiate back into a stem-like state and then differentiate into a different cell. various reprograming approaches consist of virus delivered transcription factors (1, 2), recombinant proteins (3), synthetic mrna (4), micrornas (5) and supplementation of the media with small molecules and growth factors (6).
research conducted by hochedlinger (7) and coworkers show that melanoma cells remain responsive to reprogramming into induced pluripotent stem cells (ipscs), which suggests that certain cancer cells are reprogrammable by transcription factors, which is useful for changing the epigenetic state in cancer cells. recently, kumar and coworkers showed that de-differentiation of melanoma cells is associated with the reactivation of embryonic transcription factors when they found that forced expressions of oct4 is capable of de-differentiating melanoma tumor cells to a melanoma stem cell(8).
most approaches to reprogramming pose a health risk beyond the cancer stem cells that are targeted because they involve the use of oncogenes and viruses that may integrate into the genome. this will broadly reduce the use of these methods in clinical applications and preclude fda approval. in the work described here, synthetic peptides are used instead of proteins because they are safer and easier to construct. furthermore, they can be integrated with polymeric biomaterials for potential applications in regenerative medicine.
peptide synthesis – procedures that enable the construction of materials from small peptides to large proteins (9) – enables us to create microenvironmental surfaces to which mouse melanoma cells can attach and interact with the surface bound species. we hypothesize that specific combinations of surface bound ligands will guide the epigenetic state of melanoma cells to identify a cancer stem cell. since the cellular recognition of peptide surfaces is speculated to guide intracellular signaling and downstream nuclear architecture, the cells were stained for markers of chromatin remodeling to discover possible peptide surfaces that may influence reprogramming to a melanoma stem cell state.
work by kilian and colleagues has shown that short synthetic peptides that are immobilized to self-assembled monolayers of alkanethiolates on gold can mediate the adhesion and intracellular signaling of stem cells (10). in this report, they demonstrated how the affinity and density of peptides could influence gene expression and guide lineage specification. we propose that by using adhesion peptides in combination with peptides derived from growth factors involved in reprogramming, we will be able to influence cellular epigenetics and reprogram malignant melanoma tumor cells to a melanoma stem cell.
this is a materials science approach to the matter, which focuses on the relationship between the cell and the environment around it. we hypothesize that we can apply this focus to use synthetic materials to model microenvironments around malignant cells to reprogram them into a cancer stem cell (11). this is important because it will allow us to discover the identity of the cancer stem cell, which gives us the capability to identify cancer stem cells in the body and target them with better therapeutic strategies.
2 experimental section
2.1 materials
gold-coated coverslips were used as substrate surfaces – titanium and gold were deposited by e-beam deposition onto the glass surface. all chemicals are of analytical grade and used as purchased from sigma-aldrich. chemicals consists of ether, copper-bromide, trifluoroacetic acid (tfa), n,n-dimethylformamide (dmf), methanol (meoh), dichloromethane (dcm), and diisopropylcarbodiimide (dic). tissue culture plastic ware and parafilm were used for surface preparation and cell culture. peptide synthesis reagents and amino acids were purchased from anaspec and used for solid phase peptide synthesis preparations. cell culture media and reagents were used for cell culture. mouse melanoma cells b16f0s and b16f10s (atcc) were used for cell behavior analysis. the mouse melanoma cells were used for cancer oriented research to analyze metastatic cell behavior in reaction to the microenvironment consisting of synthesized peptides.
2.2 peptide synthesis
peptides were synthesized using standard fmoc solid phase peptide synthesis methodology. n-terminal fluorenylmethyloxycarbonyl (fmoc) protected resins were deprotected in a solution with 25% piperidine and n,n-dimethylformamide (dmf) for 15 minutes. the resins were filtered and washed 4 times with dmf. three equivalents of the amino acid, benzotriazol-l-yl-oxytripyrrolidmophosphonium hexafluorophosphate (pybop), and n-methylmorpholine were dissolved in dmf and added to the resins. the solution was incubated for 1 hour at room temperature. the resins were filtered and washed 4 times with dmf and the following fmoc was deprotected. after the completion of coupling all the amino acids, the peptides were capped with a propargyl-peg-nhs ester (vendor) in dmf for 12 hours. the resins were washed 4 times with dmf and 4 times with ethanol in preparation for cleaving them from the peptide. they were cleaved with a cocktail that consists 95% trifluoroacetic acid (tfa), 2.5% h20, and 2.5% triisopropylsilane (tis) and incubated for 3 hours at room temperature. the solution was evaporated due to air flowing over it for 30 minutes. the precipitate was re-dissolved in 1 ml tfa and was re-precipitated in 9 ml of ice cold diethyl ether. the dissolve-precipitate steps were repeated an additional 3 times. the remaining materials were dissolved in water and lyophilized for 12 hours. bioactive peptides ala-thr-trp-leu-pro-pro-arg (atwlppr), his-thr-ser-asp-gln-thr-asn (htsdqtn), cys-pro-gln-pro-arg-pro-leu (cpqprpl), and ser-val-val-tyr-gly-leu-arg (svvyglr) were synthesized. the peptides were analyzed with mass spectrometry for mass purity.
2.3 surfaces
gold-plated coverslips were prepared prior to the project. they were previously cleaned by sonication in ethanol for 20 minutes. 5 nm of titanium (ti) and 20 nm of gold (au) were deposited by e-beam deposition onto the glass surface. the gold surfaces were held in dessicators for two weeks before use. when ready, the coverslips were cut into even pieces to fit into the wells of a tissue culture plate prior to cell seeding and microscopy.
2.4 cell culture
mouse melanoma cells b16f10s were cultured in high glucose dulbecco’s modified eagles mediuma containing 10% fetal bovine serum and 1% penicillin/streptomycin. 0.25% trypsin (edta) was applied to the cells to break the protein bond between the cells and the surface. they were then incubated for a few minutes while the cells separated from the surface. the cell line was passaged once and was seeded onto the surfaces at passage 3. after one hour of incubation, the cell supernatant was aspirated, the surfaces were washed, and fresh media was added.
2.5 immunohistochemistry and microscopy
the cells were fixed with paraformaldehyde and then permeabilized with triton-x 100 for 1 hour at room temperature, and blocked for one hour using 1% bovine serum albumin. the cells were immunolabeled with antibodies against acetylated lysine and nanog, and marked with fluorescently labeled secondary antibodies for microscopy. texas red-phalloidin and dapi were added to the secondary antibody solutions to label actin and nuclei respectively. the staining consisted of primary markers and secondary markers. the primary antibodies were rabbit acetylated lysine 1:500 dilution (cell signalling technologies), mouse nanog 1:500 dilution (sigma-aldrich). the secondary markers were dapi 1:5000 dilution, phalloidin texasred 1:500 dilution, anti-rabbit 488 1:500 dilution, and anti-mouse 647 1:500 dilution (secondary antibodies were purchased from life technologies and diluted in 2% goat serum). 64 (8x8 um) immunofluorescent images per well were taken by epifluorescence using an in cell analyzer 2000 (ge). images were analyzed using imagej.
3 results
3.1 peptide synthesis
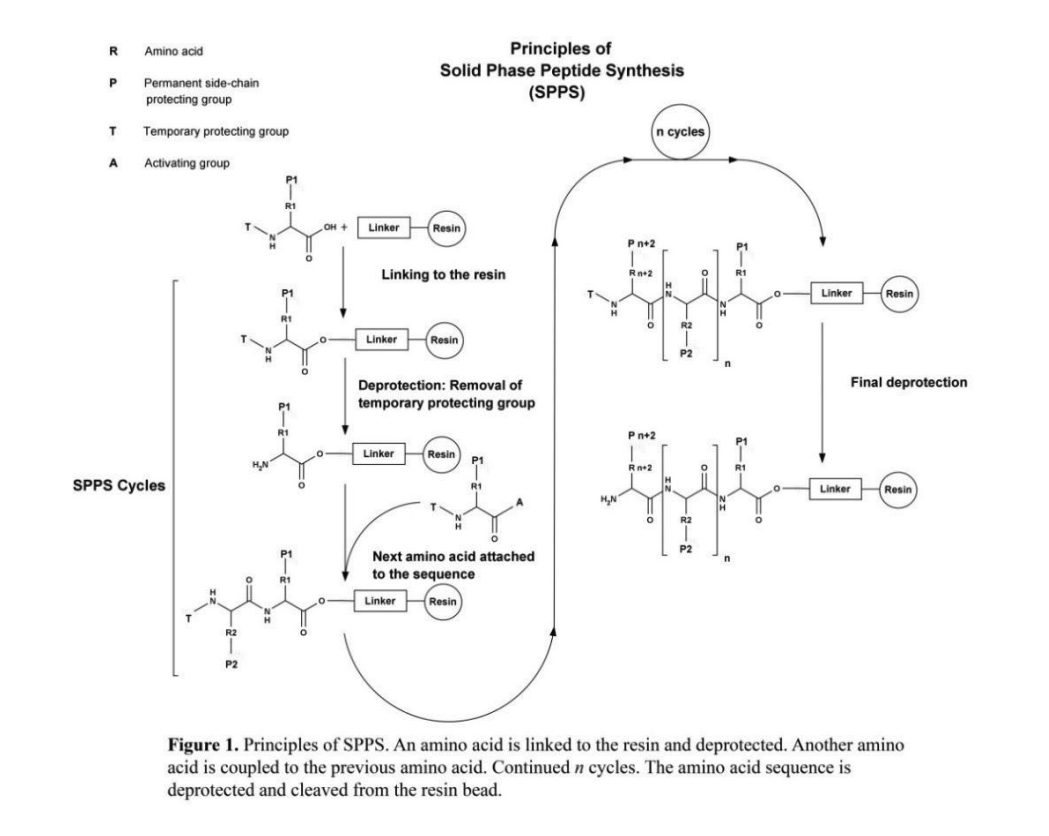
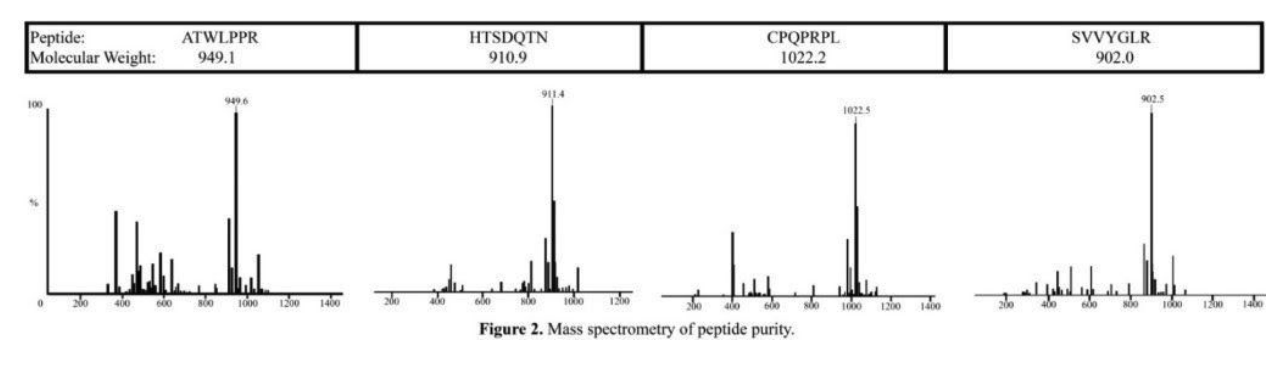
solid phase peptide synthesis (spps) – a method for manually synthesizing peptides, figure 1 shows the stepwise methodology for solid phase peptide synthesis– was used to synthesize the following peptides; atwlppr (vascular epidermal growth factor (vegf) antagonist), htsdqtn (epidermal growth factor (egf) binding peptide), cpqprpl (vegf receptor binding peptide), and svvyglr (egf receptor binding peptide). the peptides were analyzed with mass spectrometry. atwlppr was >80% purity, and htsdqtn, cpqprpl, and svvyglr were >90% purity (figure 2).
figure 1. principles of spps. an amino acid is linked to the resin and deprotected. another amino acid is coupled to the previous amino acid. continued n cycles. the amino acid sequence is deprotected and cleaved from the resin bead.
mass spectrometry of peptide purity.
3.2 sam formation
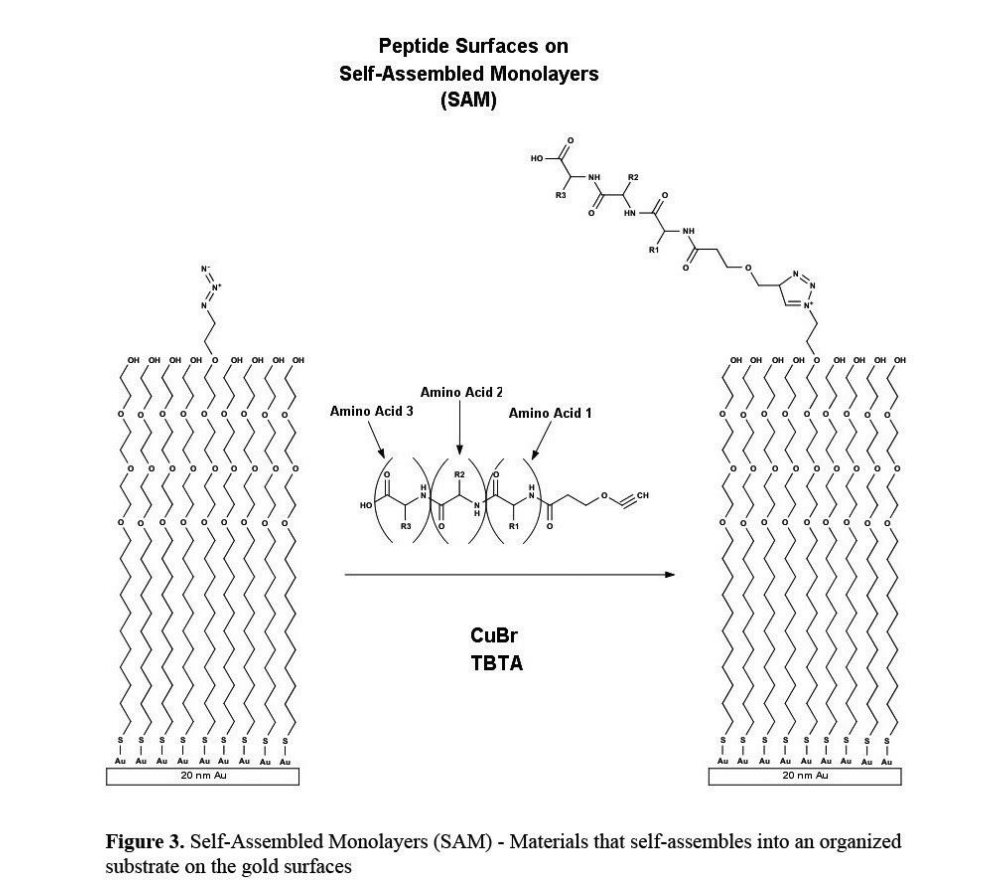
alkanethiolates are chemisorbed to the gold surface to form a self-assembled monolayer (sam) that contains a distalazide functionality for azide-alkyne cycloaddition (figure 3). peptides that were synthesized to contain an alkyne terminus were immobilized to the gold surface via “click” chemistry.
self-assembled monolayers (sam) — materials that self-assembles into an organized substrate on the gold surfaces.
3.3 cellular adhesion
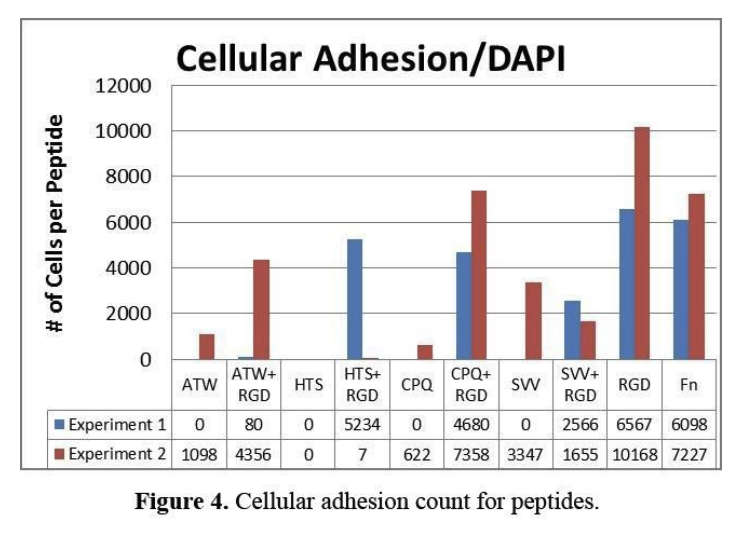
the peptides provide a surface for the cells to spread and adhere to and interact with the microenvironment. two separate experiments were performed to study the influence of the peptide surfaces on cell adhesion and the expression of reprogramming markers (figure 4). in experiment 1, it shows that there was no cellular adhesion for any of the growth factor peptides when presented alone; however the immobilized growth factor derived peptides that were mixed with an adhesion peptide did show significant numbers of cells on the surface. the cell loss on the peptides may be due to shear forces – during processing of the surfaces when immunohistochemistry was performed. in experiment 2, a higher volume solution was provided to reduce shear applied to the adherent cells and the cell count increased for most of the immobilized peptides. in both experiments the adhesion levels of fibronectin (fn) – a complex protein that mediates cell adhesion through interactions with many peptides – and rgd, a peptide derived from fn were both higher than all of the other peptides. one exception is the combination of cpq+rgd – vegf receptor binding peptide and an adhesion peptide – which showed higher cellular adhesion levels than fn in experiment 2. in both experiments, the hts peptide did not mediate cell adhesion.
cellular adhesion count for peptides.
3.4 immunofluorescent staining of melanoma cells
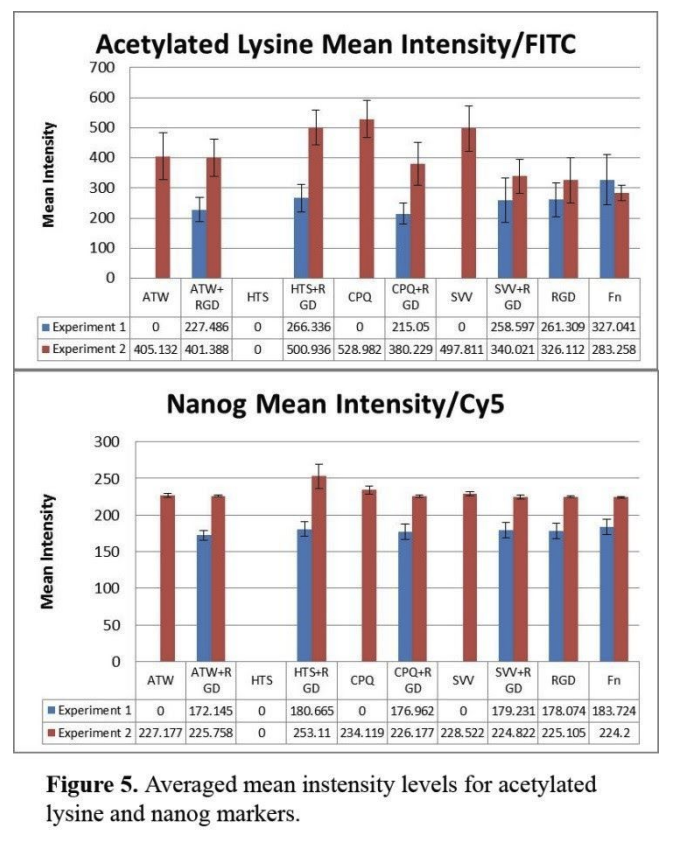
to assess whether peptide surfaces could influence cellular epigenetics, we immunostained for acetylated lysine. in experiment 1, it showed that fn was the highest average for marker expression of mean intensity acetylated lysine (figure 5). immobilized peptides, svv+rgd and hts+rgd, showed average intensity levels very similar with rgd. two other immobilized peptides, atw+rgd and cpq+rgd, showed similar intensity levels to each other and was lower than the other groups. in experiment 2, intensity levels increased substantially for all groups except fn. svv, cpq, and hts+rgd show very high levels of intensity compared to both of fn intensity levels. most immobilized peptides increased almost double the intensity in experiment 2. experiment 1 showed very similar relationships between the groups in terms of intensity levels of nanog compared to the acetylated lysine. fn had the highest averaged intensity of nanog, followed by rgd, svv+rgd, and hts+rgd at very similar levels, and then atw+rgd and cpq+rgd are lower. but cpq+rgd is closer to rgd. in experiment 2, the averaged mean intensity levels of all groups increased. hts+rgd is expressed at the highest intensity of all the groups. all the other groups are more equivalent to each other, with cpq at a slightly higher level. figure 6 shows immunofluorescent images of acetylated lysine/nanog and phalloidin-actin/nuclei for each peptide combination.
averaged mean intensity levels for acetylated lysine and nanog markers.
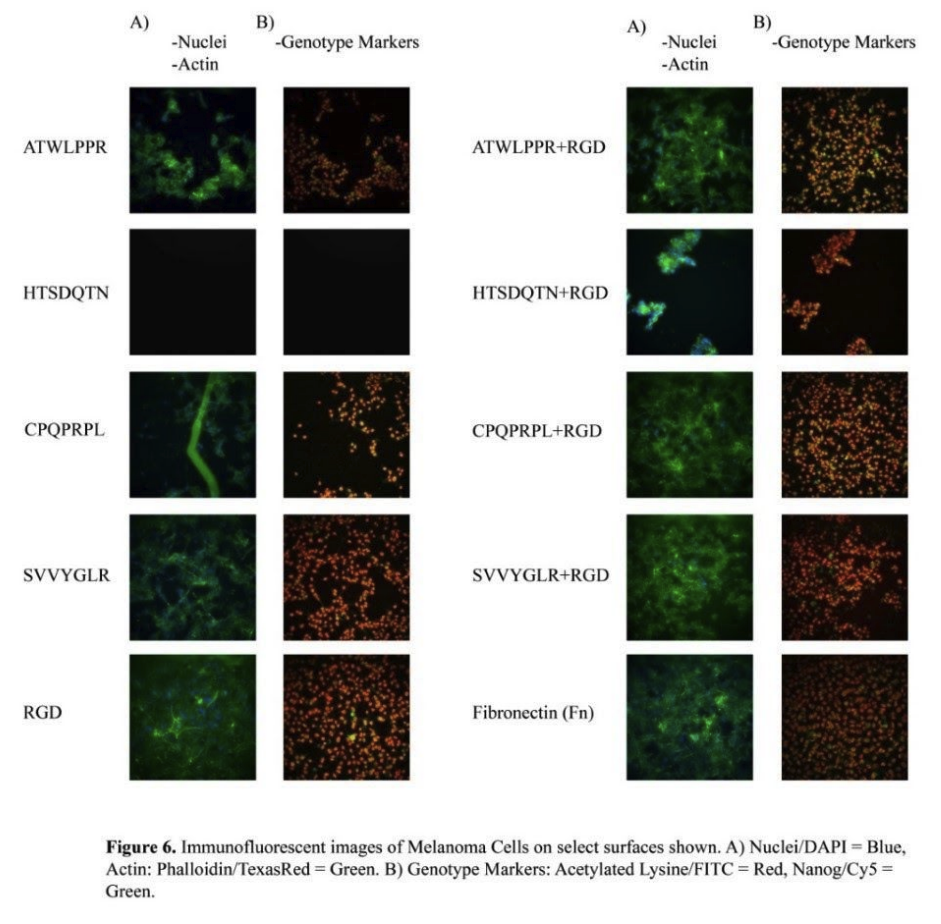
immunofluorescent images of melanoma cells on select surfaces shown. a) nuclei/dapi = blue, actin: phalloidin/texasred = green. b) genotype markers: acetylated lysine/fitc = red, nanog/cy5 = green.
4 discussion
in experiment 1, there was excessive cell loss and the conditions of the experiment were changed in experiment 2 so that there is more volume to the solution during the immunohistochemistry portion of the experiment. the increased volume allowed for the cells not to be disturbed too much while moving surfaces during immunohistochemistry. this indicates clearly that the stability of the cellular adhesion relies on conditions that do not disrupt the cells during the experiment, which requires enough volume so that cellular structures do not come off. rgd, fn, and cpq+rgd all have the highest number of cell count, which indicates that these interfaces promote the highest confluency.
when ensuring cell adhesion is unperturbed during processing (experiment 2), we see modest increases in acetylated lysine for the vegf antagonist, vegf receptor binding peptide and egf receptor binding peptide when presented alone, compared to adhesion peptide (rgd) and fibronectin. this suggests that these signaling peptides may stimulate pathways to influence the chromatin state in adherent melanoma cells. after mixing these peptides with the adhesion sequence rgd, there is a decrease in acetylation which confirms our hypothesis that these sequences promote enhanced acetylation compared to rgd alone. this shows that while rgd promotes adhesion, the hts peptide is interacting with the egf receptors and stimulating pathways that influence cellular epigenetics. while all peptides and fibronectin showed comparable levels of the pluripotency marker nanog in the adherent cells, the hts+rgd combination shows a slightly higher expression. this indicates that in the correct conditions, hts+rgd has greater intensity for nanog marker expressions than fn. this is useful because, if this genotype expression is important for reprogramming melanoma stem cells, then hts+rgd will provide a safe alternative to fn that would be approvable for therapeutic applications.
this analysis shows that different peptides have greater expression than fn for certain markers, and it also shows that it is important to recognize the extent of the peptides potential to expressing at similar levels as fn while having a majority of cells that still express at lower intensities as all of the other peptides.
5 conclusion
in this research project, the objective was to discover how mouse melanoma cells would react to adhesion peptides and combinations of cancer implicated growth factors so that it may show something interesting or useful for future applications in melanoma stem cell reprogramming for therapeutic methods in cancer research. in this project, it showed that peptides containing growth factors that enhance cancer cellular signaling shows greater potential in acetylated lysine expression than peptides that include cancer cellular signaling inhibitors. in future research and applications, these relationships between the peptide surfaces and the melanoma cells can contribute to therapeutic strategies that will be used to reprogram melanoma stem cells and identify the melanoma stem cell state.
6 references
- okita k, ichisaka t, yamanaka s. generation of germline-competent induced pluripotent stem cells. nature 2007;448(7151):313-7.
- yu j, vodyanik ma, smuga-otto k, antosiewicz-bourget j, frane jl, tian s, et al. induced pluripotent stem cell lines derived from human somatic cells. science. 2007;318:1917-20.
- zhou h, wu s, joo jy, zhu s, han dw, lin t, et al. generation of induced pluripotent stem cells using recombinant proteins. cell stem cell. 2009;4:381-4.
- warren l, manos pd, ahfeldt t, loh y-h, li h, lau f, et al. highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mrna. cell stem cell. 2010;7:618-30.
- li z, yang c-s, nakashima k, rana tm. small rna-mediated regulation of ips cell generation. embo j. 2011;30:823-34.
- yuan x, li w, ding s. small molecules in cellular reprogramming and differentiation. prog drug res. 2011;67:253-66.
- utikal j, maherali n, kulalert w, hochedlinger k. sox2 is dispensable for the reprogramming of melanocytes and melanoma cells into induced pluripotent stem cells. journal of cell science. 2009;122(19):3502-10.
- kumar sm, liu s, lu h, zhang h, zhang pj, gimotty pa, et al. acquired cancer stem cell phenotypes through oct4-mediated dedifferentiation. oncogene. 2012.
- amblard m, fehrentz ja, martinez j, subra g. methods and protocols of modern solid phase peptide synthesis. molecular biotech. 2006;33;239-53.
- kilian ka, mrksich m. directing stem cell fate by controlling the affinity and density of ligand-receptor interactions at the biomaterials interface. angew chem, int ed. 2012;51:4891-5.
- hendrix mjc, seftor ea, seftor reb, kasemeier-kulesa j, kulesa pm, postovit l-m. reprogramming metastatic tumor cells with embryonic microenvironments. nature reviews cancer. 2007;7(4):246-55.
